Article Text
Abstract
Background Cocaine use disorder (CUD) and associated psychosis are major public health issues worldwide, along with high relapse outcome and limited treatment options. Exploring the molecular mechanisms underlying cocaine-induced psychosis (CIP) could supply integrated insights for understanding the pathogenic mechanism and potential novel therapeutic targets.
Aims The aim of the study was to explore common alterations of CUD-schizophrenia-target genes and identify core risk genes contributing to CIP through data mining and network pharmacology approach.
Methods Target genes of CUD were obtained from GeneCards, Comparative Toxicogenomics Database, Swiss Target Prediction platform and PubChem. Schizophrenia-related target genes were derived from DisGeNET, GeneCards, MalaCards and Online Mendelian Inheritance in Man databases. Then, the overlap genes of these two sets were regarded as risk genes contributing to CIP. Based on these CUD-schizophrenia-target genes, functional annotation and pathway analysis were performed using the clusterProfiler package in R. Protein–protein interaction network construction and module detection were performed based on the Search Tool for the Retrieval of Interacting Genes (STRING) database and Cytoscape software. Gene expression datasets GSE54839 and GSE93577 were applied for data validation and diagnostic capacity evaluation of interested hub genes.
Results A total of 165 CUD-schizophrenia-target genes were obtained. These genes were mainly contributing to chemical synaptic transmission, neuropeptide hormone activity, postsynaptic membrane and neuroactive ligand–receptor interaction pathway. Network analysis and validation analysis indicated that BDNF might serve as an important risk gene in mediating CIP.
Conclusions This study generates a holistic view of CIP and provides a basis for the identification of potential CUD-schizophrenia-target genes involved in the development of CIP. The abnormal expression of BDNF would be a candidate therapeutic target underlying the pathogenesis of CUD and associated CIP.
- schizophrenia
- psychiatry
Data availability statement
Data are available on reasonable request. All data and codes are available from the corresponding author or first author by request. The microarray dataset used in the paper can be downloaded from the GEO database.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Introduction
Cocaine is one of the most prevalent illicit drugs worldwide, and cocaine use disorder (CUD) has become a major public health concern. Moreover, the overall mortality is substantially elevated in patients who use cocaine. The Global Burden of Disease study estimated that the age-adjusted prevalence of CUD was 64 per 100 000 population, especially in high-income countries.1 Beyond the rising mortality, chronic cocaine abuse and CUD were also associated with significant adverse abnormalities in brain function,2 leading to numbers of negative consequences including cognitive impairments, high impulsivity as well as aggressive behaviours, poor psychological well-being and even morbidity of other mental disorders.3 Psychotic symptoms and syndromes are among the common consequences of abusers, with approximately 40% of people affected.4 The prevalence of psychiatric-related symptoms is 11 times higher compared with the general population.5 Common symptoms include hallucinations, delusions, agitation, violence, irritability, anxiety as well as depression.6 Although acute and transient psychotic symptoms account for the largest proportion, there are a considerable number of patients who gradually develop into persistent related mental disorders. The long-lasting disorders lead to a greater disease and mental health burden, resulting in progressive social and occupational deterioration.7
Genetic risks are important aetiological factors that give rise to psychotic symptoms and syndromes among these patients.8 Moreover, cocaine-related psychiatric symptoms are quite similar to schizophrenia in terms of symptomatology and pathogenesis.9 In the meantime, individuals with schizophrenia spectrum disorders showed much heightened risk of substance abuse and exacerbating pre-existing psychosis.10 This evidence indicates that these two disorders might share predisposing genetic factors and neural signalling pathways. Several psychiatric-related overlap genes have been identified as potential mediators of cocaine-induced psychosis (CIP) and schizophrenia. Genetic variations of glutamate ionotropic receptor NMDA type subunit 1 (GRIN1), solute carrier family 6 member 4 (SLC6A4), dystrobrevin binding protein 1 (DTNBP1) and superoxide dismutase 2 (SOD2)11 have been found to be significantly associated with both CUD resultant psychosis and schizophrenia. Experimental studies have shown that CIP symptoms correlate with dopaminergic and GABAergic6 mechanisms. Respectively, dopamine and gamma-aminobutyric acid (GABA)-related pathways were important mediators contributing to the symptomatology and pathology of schizophrenia. Taken together, understanding the affected pathways and specific gene expression profiles may help better uncovering disease-associated biomarkers for clinical conceptualisation and personalisation treatment of CUD and related psychosis. However, contrasting results have been observed in different gene expression studies and there is still lack of significant biomarkers of CIP. One possible reason is that substance use disorder is a disease involving multiple brain areas, including prefrontal cortex, midbrain limbic system, nucleus accumbens and ventral tegmental area. Second, gene expression profiles of peripheral cells may be different from postmortem brain tissues.
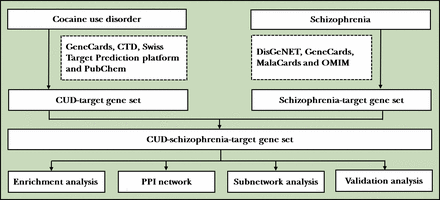
This problem can be partly solved by integrated analysis of gene expression profiles from multiple datasets, online databases and network pharmacology approach. The rapidly developing bioinformatics and data mining technologies have been widely applied to identify psychiatric disease-associated biomarkers that contribute to high-risk individual screening, diagnosis, classification, prognostic and recurrence prediction. On this basis, the aim of this study was to elucidate overlap gene expression changes in the pathogenesis of CIP and schizophrenia, in order to acquire new potential biomarkers for diagnostic and medication of these CUD and related psychiatry disorders accurately (figure 1).
Flowchart of the study. CTD, Comparative Toxicogenomics Database; CUD, cocaine use disorder; OMIM, Online Mendelian Inheritance in Man; PPI, protein–protein interaction.
Materials and methods
Collection of potential CUD-schizophrenia-target gene set
For putative target genes of CUD, ‘cocaine’, ‘CUD’, ‘cocaine addiction’ and ‘cocaine dependence’ were used as keywords to query the following electronic databases: GeneCards (https://www.genecards.org/), Comparative Toxicogenomics Database (CTD; http://ctdbase.org/), Swiss Target Prediction platform (http://www.swisstargetprediction.ch/) and PubChem. For schizophrenia-related target genes, disease-related online database DisGeNET (https://www.disgenet.org/), GeneCards (https://www.genecards.org/), MalaCards (https://www.malacards.org/) and Online Mendelian Inheritance in Man (OMIM, https://omim.org/) databases were applied for target screening. Then, after removing the duplicates, a CUD-target gene set (gene set 1) as well as a schizophrenia-target gene set (gene set 2) was established by intersecting the search results of four databases, respectively. Finally, CUD-schizophrenia-target gene set was obtained by the intersection of gene set 1 and gene set 2. The overlapping genes of these two gene sets were visualised using VennDiagram package in R (V.3.6.3).
PPI network construction and module identification
Protein–protein interaction (PPI) networks of differently expressed genes (DEGs) were analysed using the Search Tool for the Retrieval of Interacting Genes (STRING) database (V.9.1, https://string-db.org). Combined score >0.7 (high confidence) was considered as calculation criterion. Two Cytoscape plug-ins were applied to screen the core genes and critical subnetwork. First, the most significant modules of PPI network were screened by the Molecular Complex Detection (MCODE). MCODE scores>5, node score cut-off=0.2, maximum depth=100 and k-score=2 were set as filter criteria. Second, CytoNCA plug-in of Cytoscape was used for calculating the scores of each node. Genes with node scores higher than the median value of betweenness, closeness and degree were considered as core genes. Network and subnetwork visualisation was carried out through Cytoscape (V.3.7.2).
Functional enrichment analysis of CUD-schizophrenia-target gene set
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of CUD-schizophrenia-target genes were implemented through clusterProfiler package in R (V.3.6.3). Criterion was set as p value <0.05 and false discovery rate <0.05.
Microarray data validation
To verify the robustness of hub genes, human microarray expression data files focusing on cocaine-related illicit drugs and schizophrenia were searched in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). The inclusion criteria were as follows: (1) the series type in GEO was expression profiling by array, (2) the organism was Homo sapiens, (3) the samples should include patients and controls and (4) clinical tissues (blood or brain) must be contained. Exclusion criteria were as follows: (1) microarray samples were cell lines or animal models, (2) dataset did not contain original data and (3) dataset did not provide any demographic data of array samples. GEO datasets meeting these criteria were enrolled for further analysis.
Results
CUD-schizophrenia-target gene set acquisition and screening
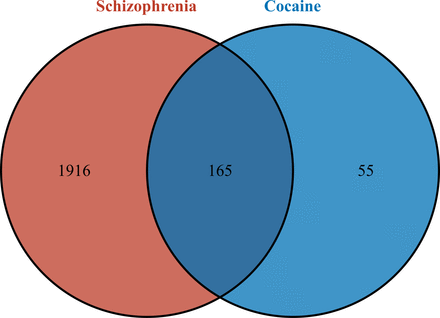
A total of 220 cocaine-related target genes and 2081 schizophrenia-related target genes were obtained through database prediction and literature research. By intersecting the two sets, a total of 165 CUD-schizophrenia-target genes were obtained (figure 2).
Identification of CUD-schizophrenia-target genes. CUD, cocaine use disorder.
PPI network construction and module identification
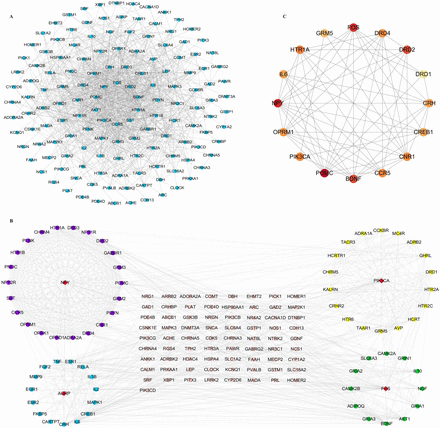
PPI network includes 144 nodes and 879 gene–gene interaction edges (figure 3A). Module analysis revealed four most significant modules (figure 3B), with NPY, PIK3CA, AGRP and FOS serving as seed genes for each module, respectively. In CytoNCA network analysis, 16 target nodes and 63 target–target interactions were found (figure 3C). POMC, FOS, NPY, BDNF and DRD2 showed the highest degree, betweenness and closeness scores (table 1).
(A) PPI network of 165 selected CUD-schizophrenia-related genes. The nodes with the highest degree are in the inner circle. (B) The most significant modules in PPI network. Red diamond nodes are representative of seed genes. Modes marked in purple belong to module 1, and NPY genes are identified as seed genes. Module 2 is marked in yellow; the seed gene is PIK3CA. In module 3, all nodes are shown in blue circle, with AGRP serving as seed gene. For module 4, related nodes are marked in green. Their seed gene is FOS. (C) Subnetwork of the CytoNCA. The shade of the colour indicates the score of the node. POMC and FOS show the highest score. CUD, cocaine use disorder; PPI, protein–protein interaction.
CytoNCA network scores of screened genes
Functional enrichment analysis of CUD-schizophrenia-target gene set
GO enrichment analysis showed that changes in biological processes (BPs) were mainly enriched in chemical synaptic transmission (GO:0007268), behavioural response to cocaine (GO:0048148) and response to drug (GO:0042493), while in molecular function (MF) ontology, these genes were significantly enriched in neuropeptide hormone activity (GO:0005184), drug binding (GO:0008144) and kinase activity (GO:0016301). For changes in cellular component (CC) ontology, postsynaptic membrane (GO:0045211), postsynaptic density (GO:0014069) and cell junction (GO:0030054) were the most critical CC terms. In KEGG pathway analysis, the most enriched pathways were neuroactive ligand–receptor interaction (hsa04080), cyclic adenosine monophosphate (cAMP) signalling pathway (hsa04024) and amphetamine addiction pathway (hsa05031). The top 10 most significant enriched GO and KEGG terms of CUD-schizophrenia-related genes are illustrated in figure 4.
Enrichment analysis. Top 10 enriched GO and KEGG terms of CUD-schizophrenia-related genes. Gene ratio refers to the ratio of enriched genes compared with all 165 target genes. Gene count refers to the number of enriched genes. The size of the bubbles is on behalf of target gene concentration of the significant enriched items. The colour depth of the bubble is representative of significance. BP, biological process; cAMP, cyclic adenosine monophosphate; CC, cellular component; CUD, cocaine use disorder; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function; TNF, tumor necrosis factor.
Microarray data validation
In the end, two eligible microarray databases were selected for validation. GSE54839 is a cocaine addiction-related microarray based on GPL6947 platform. GSE54839 dataset contains gene expression profiles from 10 midbrain postmortem samples of chronic cocaine abusers and 10 well-matched drug-free control subjects.12 These subjects were all men. The average age of cocaine group was 50.10 (4.51) years and the average age of control group was 48.30 (3.09) years. No significant differences in data between the two groups were observed (p=0.312). GSE93577 was based on the GPL13667 platform. In GSE93577, prefrontal cortex tissues which were derived from 36 well-matched pairs of patients with schizophrenia and unaffected health subjects were included for validation analysis. Average ages were 48.1 (13.0) years and 46.9 (12.4) years for patients with schizophrenia and control subjects, respectively. There were also no significant differences in sex (p=1.000), race (p=0.100) and age (p=0.690) between the two groups.13
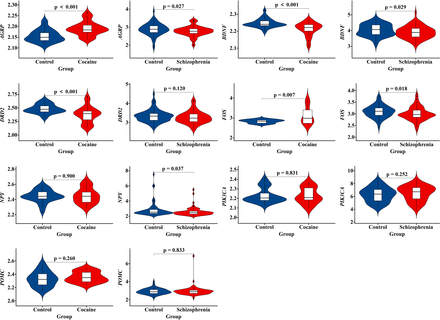
Top five genes with the highest scores (POMC, FOS, NPY, BDNF and DRD2) and four seed genes (NPY, FOS, AGRP and PIK3CA) in the most significant modules were selected for validation analysis. To overcome the limitations of individual studies, we validated our findings in datasets GSE54839 and GSE93577. Results showed that BDNF, FOS and AGRP showed significant expression difference both in schizophrenia dataset GSE93577 and cocaine addiction dataset GSE54839. However, only BDNF showed a consistent downregulated expression trend in both selected datasets. On the contrary, FOS and AGRP demonstrated a downregulated expression trend in schizophrenia dataset GSE93577 and an upregulated expression trend in cocaine addiction dataset GSE54839 (figure 5).
Violin plot of expression levels of the top identified cocaine-schizophrenia-target genes. Genes that reached significant different expression levels were FOS, AGRP and BDNF. Only BDNF showed a consistent downregulated expression trend in both schizophrenia dataset GSE93577 and cocaine addiction dataset GSE54839. FOS and AGRP demonstrated a downregulated expression trend in schizophrenia dataset GSE93577 and an upregulated expression trend in cocaine addiction dataset GSE54839.
Discussion
Main findings
Gene polymorphism is recognised as one of the crucial factors leading to substance-induced psychosis. However, the exact molecular mechanisms of gene mutations remain poorly understood. In addition, owing to the lack of a therapeutic target, CUD is still a disease with high relapse rate, imposing a considerable burden on the whole society. Therefore, identifying the susceptibility genes and key mediators is imperative to develop novel treatment approaches for CUD and CIP. In this study, a total of 165 CUD-schizophrenia-target genes were obtained. These risk genes were mainly contributing to chemical synaptic transmission (Gene Ontology: BP), neuropeptide hormone activity (Gene Ontology: MF), postsynaptic membrane (Gene Ontology: CC) and neuroactive ligand–receptor interaction (KEGG pathway). Network analysis and validation analysis indicated that BDNF might serve as a key gene mediating CIP. Compared with FOS and AGRP, which showed reversed trends in microarray data GSE54839 and GSE93577, BDNF was the only significant downregulated gene in postmortem brain samples of cocaine addiction and schizophrenia brain tissues.
Brain-derived neurotrophic factor (BDNF) is a protein the coding gene of which locates in chromosome 11. BDNF affects neural development and synaptic plasticity associated with many common psychiatric disorders and has long been recognised to play a major role in learning and memory processes. Through the combination of its high affinity receptor of tropomyosin receptor kinase B (trkB), BDNF signalling cascades were activated to affect gene transcription and synaptic structure and plasticity.14
BDNF is closely involved in cocaine-related behaviours, such as craving, drug seeking and decision making.15 Research has indicated that reduced BDNF signalling appears to increase the risk of stimulant addiction development16 and might serve as a reliable biomarker for cocaine addiction.17 Repeated administration of cocaine leads to a reduction of BDNF expression in multiple brain areas. In the prefrontal cortex, cocaine administration could decrease BDNF levels and synaptic density, affecting cortical–striatal glutamatergic signalling.18 These anomalies can further result in cocaine-related signal restoring and drug-seeking behavioural alterations.19 Repeated administration of cocaine also leads to a reduction of BDNF level in hippocampus, resulting in altered dendritic and spine morphology.20 The maintenance of drug-related midbrain dopaminergic and cholinergic neurons was also highly related to cocaine administration.21
BDNF also played a crucial role in schizophrenia. The abnormal BDNF signalling can lead to abnormal brain functioning, making patients more susceptible to schizophrenia and relapse.22 Decreased BDNF has already been confirmed in patients with schizophrenia and animal models.23 In addition, BDNF can effectively penetrate the blood–brain barrier in both directions. Central BDNF levels were positively correlated with peripheral BDNF levels,24 which make BDNF an optimal risk gene for both CIP and schizophrenia.
BDNF is also in close relationship with CUD and CIP medication. Our network analysis and validation study reveal that BDNF levels were lower than control samples in both patients who use cocaine and patients with schizophrenia. As a neurotrophin, BDNF plays an important role in modulating dopaminergic neuron differentiation and establishment, affecting dopaminergic and serotonergic neurotransmission.25 These were all crucial pathways contributing to the pathogenesis and pathophysiology of schizophrenia and cocaine addiction.26 In relation to CIP, BDNF level was decreased in patients with CIP, which suggested that BDNF might play a role in the psychotic symptoms associated with cocaine consumption.27 Increased BDNF was strongly associated with cocaine abstinence treatment compliance.28 Previous studies have provided preliminary evidence that regulating BDNF-trkB binding could partially suppress cocaine-induced drug-seeking behaviour.29 In addition, the polymorphism of the BDNF val66met was proved to be a potential pharmacogenetic target gene for substance addiction and related disorders.30 Taken together, these combined evidence suggested that BDNF, which showed the co-directional expression trend in both cocaine and schizophrenia samples, might serve as a key risk gene mediating cocaine addiction and CIP.
Limitations
There are several limitations in our study. First, human microarray studies on psychiatry and substance use disorders were often associated with small sample sizes and led to limited statistical power. Second, in a validation study, microarray dataset GSE54839 was analysed in postmortem midbrain tissues, which might not entirely reflect gene expression changes in peripheral and other brain areas. Third, CIP risk gene screening was only based on prediction and validation from public databases, and further in vivo and in vitro confirmatory experiments are needed to confirm the results of our research.
Implications
Understanding the most affected pathways and specific gene expression profiles in the pathogenesis of CIP may help to uncover disease-associated biomarkers for risk assessment, regulatory mechanisms and personalisation of treatment. In our study, 165 CUD-schizophrenia-target genes were identified as candidate targets for CIP. BDNF would be a valuable risk gene or potential therapeutic target contributing to the pathogenesis of CUD and related psychosis.
Data availability statement
Data are available on reasonable request. All data and codes are available from the corresponding author or first author by request. The microarray dataset used in the paper can be downloaded from the GEO database.
Ethics statements
Patient consent for publication
Ethics approval
The study protocol was approved by the institutional review board at the Shanghai Mental Health Centre (approval certificate number: 2016-11R). All procedures followed were in accordance with the ethical standards of the Norwegian National Committee for Research Ethics in the Social Sciences and the Humanities and with the Helsinki Declaration of 1975, as revised in 2000.
Acknowledgments
The authors would like to thank the research teams that uploaded the microarray datasets GSE54839 and GSE93577 to GEO database.
References
Zhu Youwei obtained a PhD degree in psychiatry from Shanghai Jiao Tong University, China, in 2018. At present, he is working as a public health doctor at the Shanghai Mental Health Center in China. His main research interests include vocational rehabilitation of severe mental disorder, cognitive rehabilitation treatment and substance use disorder.

Footnotes
Contributors YouZ and YanZ are responsible for the statistical analysis and the writing of the original paper. XX, HS and XL are responsible for database and literature researching of bioingredients and disease targets. HJ is responsible for crosschecking the screened data and quality control. English language editing of the paper was also carried out by JD and NZ. MZ is in charge of the whole research and is responsible for the scientific design of this study.
Funding This work was supported by the National Nature Science Foundation (81771436), Program of Shanghai Academic Research Leader (17XD1403300), Shanghai Municipal Health and Family Planning Commission (2017ZZ02021, 2018YQ45 and 20184Y0134), Shanghai Key Laboratory of Psychotic Disorders (13DZ2260500), Shanghai Municipal Science and Technology Major Project (2018SHZDZX05), Shanghai Sailing Program (19YF1442100) and Shanghai Mental Health Center Program (2018-QH-02, 2020-YJ-06, CRC2018YB02).
Disclaimer The funders have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests None declared.
Provenance and peer review Not commissioned; externally peer reviewed.